I’ve got PyData NYC 2018 in two days and rather finishing up my talk, I just realized that my source data has a silent corruption due to non-standard timestamps. Here’s how I fixed this using pandas and then uploaded the data to OmniSci.
Computers Are Dumb, MAKE THINGS EASIER FOR THEM!
Literally every data tool in the world can read the ISO-8601 timestamp format. Conversely, not every tool in the world can read Excel or whatever horrible other tool people use to generate the CSV files seen in the wild. While I should’ve been more diligent checking my data ingestion, I didn’t until I created a wonky report…
Let’s take a look at the format that tripped me up:
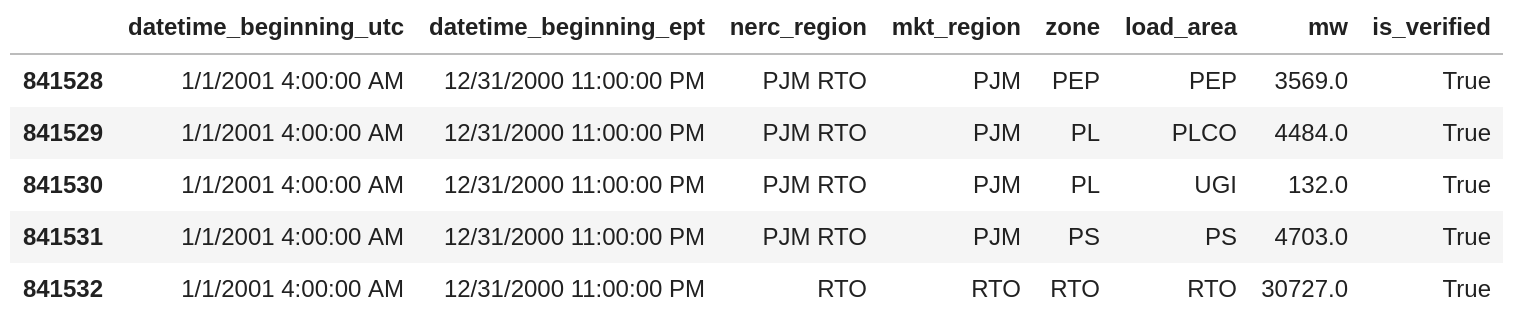
Month/Day/Year Hour:Minute:Second AM/PM feels very much like an Excel date format that you get when Excel is used as a display medium. Unfortunately, when you write CSV files like this, the next tool to read them has to understand 1) that these columns are timestamps and 2) if the user doesn’t specify the format, has to guess the format.
In my case, I didn’t do descriptive statistics on my timestamp columns and had a silent truncation(!) of the AM/PM portion of the data. So instead of having 24 hours in the day, the parser read the data as follows (the #AM and #PM are my comments for clarity):
1 | |
So while the data looks like it was imported correctly (because, it is a timestamp), it wasn’t until I realized that hours 13-23 were missing from my data that I realized I had an error.
Pandas To The Rescue!
Fixing this issue is as straight-forward as reading the CSV into python using pandas and specifying the date format:
1 | |
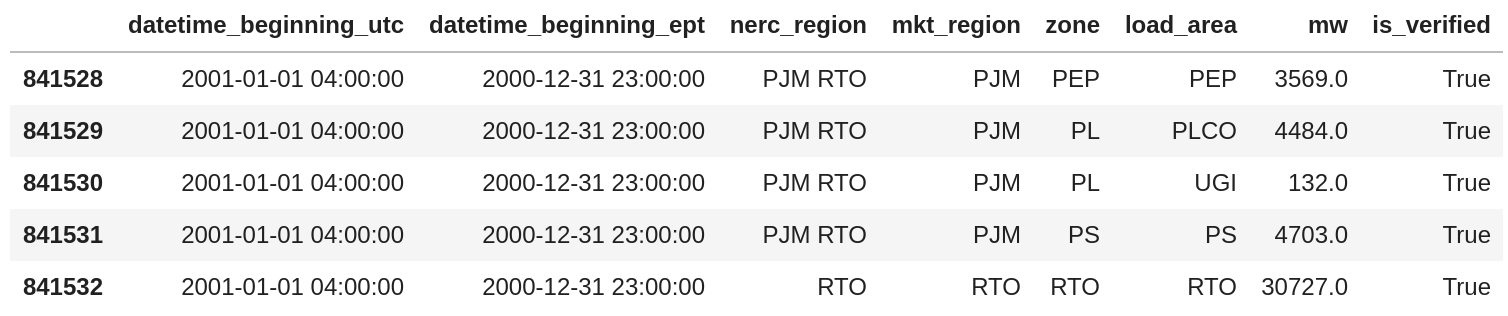
We can see from the code above that pandas has taken our directive about the format and it appears the data have been parsed correctly. A good secondary check here is that the difference in timestamps is -5, which is the offset of the East Coast of the United States relative to UTC.
Uploading to OmniSci Directly From Pandas
Since my PyData talk is going to be using OmniSci, I need to upload this corrected data or rebuild all my work (I’ll opt for fixing my source). Luckily, the pymapd package provides tight integration to an OmniSci database, providing a means of uploading the data directly from a pandas dataframe:
1 | |
I have a pre-existing table hrl_load_metered on the database, so I can truncate the table to remove its (incorrect) data but keep the table structure. Then I can use load_table_columnar to insert the cleaned up data into my table and now my data is correct.
Computers May Be Dumb, But Humans Are Lazy
At the beginning, I joked that computers are dumb. Computers are just tools that do exactly what a human programs them to do, and really, it was my laziness that caused this data error. Luckily, I did catch this before my talk and the fix is pretty easy.
I’d like to say I’m going to remember to check my data going forward, but in reality, I’m just documenting this here for the next time I make the same, lazy mistake.