I’m a postdoc studying scientific reproducibility. I have a machine learning question that I desperately need your help with. My advisor and I disagree on how we should carry out repeated cross-validation. We would love to have a third expert opinion…
I’m trying to predict whether a study can be successfully replicated (DV), from the texts in the original published article. Our hypothesis is that language contains useful signals in distinguishing reproducible findings from irreproducible ones. The nuances might be blind to human eyes, but can be detected by machine algorithms.
The protocol is illustrated in the following diagram to demonstrate the flow of cross-validation. We conducted a repeated three-fold cross-validation on the data.
STEP 1) Train a doc2vec model on the training data (2/3 of the data) to convert raw texts into vectors representing language features (this algorithm is non-deterministic, the models and the outputs can be different even with the same input and parameter)STEP 2) Infer vectors using the doc2vec model for both training and test setsSTEP 3) Train a logistic regression using the training setSTEP 4) Apply the logistic regression to the test set, generate a predicted probability of success
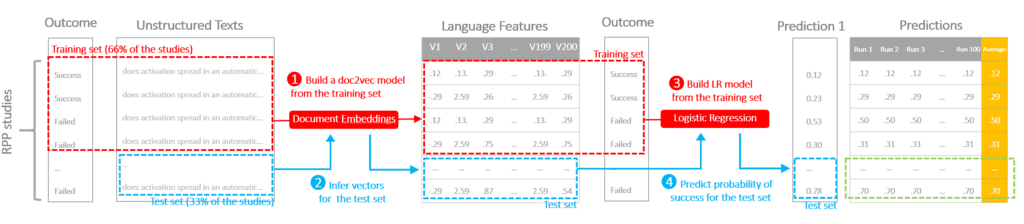
Because doc2vec is not deterministic, and we have a small training sample, we came up with two choices of strategies:
(1) All studies were first divided into three subsamples A, B, and C. Step 1 through 4 was done once with sample A as the test set, and a combined sample of B and C as the training set, generating on predicted probability for each study in sample A. To generate probabilities for the entire sample, Step 1 through 4 was repeated two more times, setting sample B or C as the test set respectively. At this moment, we had one predicted probability for each study. Subsequently, the entire sample was shuffled to create a different random three-fold partition, followed by same three-fold cross-validation. A new probability was generated for each study this time. The whole procedure was iterated 100 times, so each study had 100 different probabilities. We averaged the probabilities and compared the average probabilities with the ground truth to generate a single AUC score.
(2) All studies were first divided into three subsamples A, B, and C. Step 1 through 4 was first repeated 100 times with sample A as the test set, and a combined sample of B and C as the training set, generating 100 predicted probabilities for each study in sample A. As I said, these 100 probabilities are different because doc2vec isn’t deterministic. We took the average of these probabilities and treated that as our final estimate for the studies. To generate average probabilities for the entire sample, each group of 100 runs was repeated two more times, setting sample B or C as the test set respectively. An AUC was calculated upon completion, between the ground truth and the average probabilities. Subsequently, the entire sample was shuffled to create a different random three-fold partition, followed by the same 3×100 runs of modeling, generating a new AUC. The whole procedure was iterated on 100 different shuffles, and an AUC score was calculated each time. We ended up having a distribution of 100 AUC scores.
I personally thought strategy two is better because it separates variation in accuracy due to sampling from the non-determinism of doc2vec. My advisor thought strategy one is better because it’s less computationally intensive and produce better results, and doesn’t have obvious flaws.