** Tue 03 January 2017
There have been many Python libraries developed for interacting with the Hadoop File System, HDFS, via its WebHDFS gateway as well as its native Protocol Buffers-based RPC interface. I’ll give you an overview of what’s out there and show some engineering I’ve been doing to offer a high performance HDFS interface within the developing Arrow ecosystem.
This blog is a follow up to my 2017 Roadmap post.
Hadoop file system protocols
HDFS is a part of Apache Hadoop, and its design was originally based on the Google File System described in the original MapReduce paper. Its native wire protocol uses’s Google Protocol Buffers (or “protobufs” for short) for remote procedure calls, or RPCs.
Traditionally, systems that talk to HDFS, like the main Java client library, would implement the Protobuf messaging format and RPC protocol. To make it easier for light load applications to read and write files, WebHDFS was developed to provide an HTTP or HTTPS gateway to make PUT and GET requests instead of protobuf RPCs.
For light load applications, WebHDFS and native protobuf RPCs provide comparable data throughput, but native connectivity is generally considered to be more scalable and suitable for production use.
Python has two WebHDFS interfaces that I’ve used:
The rest of this article will focus instead on native RPC client interfaces.
Native RPC access in Python
The “official” way in Apache Hadoop to connect natively to HDFS from a C-friendly language like Python is to use libhdfs, a JNI-based C wrapper for the HDFS Java client. A primary benefit of libhdfs is that it is distributed and supported by major Hadoop vendors, and it’s a part of the Apache Hadoop project. A downside is that it uses JNI (spawning a JVM within a Python process) and requires a complete Hadoop Java distribution on the client side. Some clients find this unpalatable and don’t necessarily require the production-level support that other applications require. For example, Apache Impala (incubating), a C++ application, uses libhdfs to access data in HDFS.
Due to the heavier-weight nature of libhdfs, alternate native interfaces to HDFS have been developed.
libhdfs3, now part of Apache HAWQ (incubating), a pure C++ library developed by Pivotal Labs for use in the HAWQ SQL-on-Hadoop system. Conveniently, libhdfs3 is very nearly interchangeable for libhdfs at the C API level. At one time it seemed that libhdfs3 might become a part of Apache Hadoop officially, but that does not now seem likely (see HDFS-8707, a new C++ client in development).
snakebite: a pure Python implementation of Hadoop’s protobuf RPC interface, created by Spotify.
Since snakebite does not offer a comprehensive client API (e.g. it cannot write files) and has worse performance (being implemented in pure Python), I’ll focus on libhdfs and libhdfs3 going forward.
Python interfaces to libhdfs and libhdfs3
There have been a number of prior efforts to build C-level interfaces to the
libhdfs JNI library. These includes cyhdfs (using Cython), libpyhdfs
(plain Python C extension), and pyhdfs (using SWIG). One of the challenges
with building a C extension to libhdfs is that the libhdfs.so shared library
is distributed with the Hadoop distribution, so you must properly configure the
$LD_LIBRARY_PATH so that the shared library can be loaded. Additionally, the
JVM’s libjvm.so must also be loaded at import time. Combined, these lead to
some “configuration hell”.
When looking to build a C++ HDFS interface for use in Apache Arrow (and Python via PyArrow), I discovered the libhdfs implementation in Turi’s SFrame project. This takes the clever approach of discovering and loading both the JVM and libhdfs libraries at runtime. I adapted this approach for use in Arrow, and it has worked out nicely. This implementation provides very low-overhead IO to Arrow data serialization tools (like Apache Parquet), and convenient Python file interface.
Because the libhdfs and libhdfs3 driver libraries have very nearly the same C API, we can switch between one driver and the other with a keyword argument in Python:
In parallel, the Dask project developers created hdfs3, a pure Python interface to libhdfs3 that uses ctypes to avoid C extensions. It provides a Python file interface and access to the rest of the libhdfs3 functionality:
pyarrow.HdfsClient and hdfs3 data access performance
Running against a local CDH 5.6.0 HDFS cluster, I computed ensemble average performance in a set of file reads of various sizes from 4 KB to 100 MB under 3 configurations:
-
hdfs3(which always uses libhdfs3) -
pyarrow.HdfsClientusingdriver='libhdfs' -
pyarrow.HdfsClientusingdriver='libhdfs3'
You can obtain all of these packages by running:
Note: pyarrow conda-forge packages are currently only available for Linux; in theory this will be resolved by January 20, 2017. If anyone would like to help with Windows support, let me know.
Performance numbers are in megabytes per second (“throughput”). Benchmarking code is shown at the end of the post. I am very curious about results in more diverse production environments and Hadoop configurations.
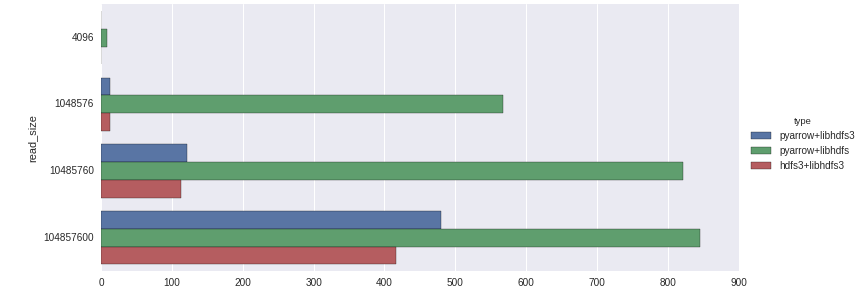
Curiously, at least in my testing, I found these results:
libhdfs, despite being Java and JNI-based, achieves the best throughput in this test. libhdfs3 performs poorly in small size reads. This may be due to some RPC latency or configuration issues that I am not aware of. Strictly comparing libhdfs3, pyarrow outperforms hdfs3 by a 10-15% margin; I expect this is primarily due to memory handling / copying based on the ctypes (hdfs3) vs. C++ (pyarrow) implementation.
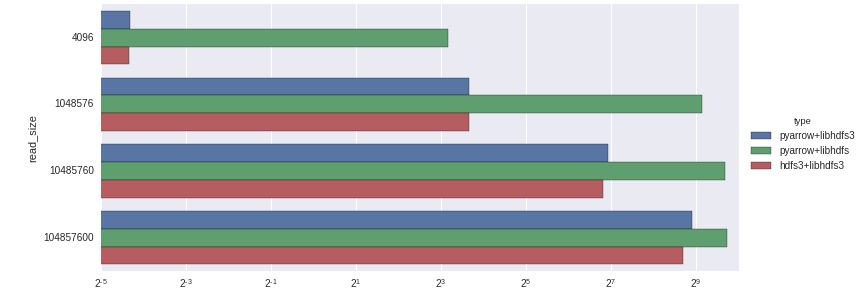
Here are the timings with a logarithmic axis:
Native C++ IO in Apache Arrow
One of the reasons for bunding IO interfaces like HDFS in the pyarrow library is that they all utilize a common memory management layer that enables data to be passed around with very low (and sometimes zero) copying overhead. By comparison, libraries that expose only a Python file interface introduce some amount of overhead because memory is being handled by bytes objects in the Python interpreter.
Full details of the Arrow’s C++ IO system are out of the scope of this article, but I’ll write a future blog post taking a deeper dive into the details.