** Mon 22 February 2016
I’m super excited to be involved in the new open source Apache Arrow community initiative. For Python (and R, too!), it will help enable
- Substantially improved data access speeds
Closer to native performance Python extensions for big data systems like Apache Spark
- New in-memory analytics functionality for nested / JSON-like data
There’s plenty of places you can learn more about Arrow, but this post is about how it’s specifically relevant to pandas users. See, for example:
Accelerating data access for pandas users on Hadoop clusters
For average pandas users, the gold standard for storing and retrieving data on local machines (or network file systems) is usually one of:
-
CSV files, using
pandas.read_csv -
HDF5 data format files, using
pandas.HDFStore -
Another binary dataformat, like the Blosc-powered bcolz
But if your data is in a Hadoop cluster, it may not be as simple as reading a file off disk. Here’s some of the data-providing systems and storage formats you can access from pandas:
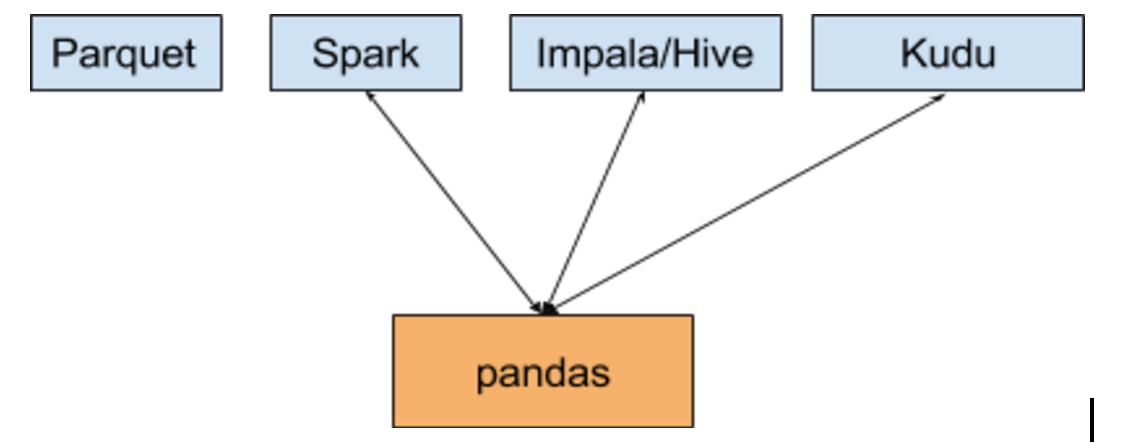
Unfortunately, the quality of these data connections for pandas are highly
variable. I did an in-depth exploration to compare the performance of
retrieving a pandas.DataFrame with 1 million rows with a net footprint of
about 90 megabytes. Here is the performance summary:
|Method|Speed (sec)|vs HDF5|vs read_csv|Effective speed
|pandas.HDF5Store (uncompressed)|0.117|1.0x|0.05x|769.23 MB/s|
|Spark DataFrame.toPandas|13.6|116.2x|5.60x|6.62 MB/s|
|Impala SELECT (via impyla)|15.3|130.8x|6.30x|5.88 MB/s|
As the original author of both read_csv and HDFStore, these benchmarks make
me very proud, but it also shows what level of data access performance pandas
users should expect in general.
The price of data serialization can be directly seen in the Impala query profile (note, this is a DEBUG Impala build):
So more than 90% of the execution time is data serialization.
The reasons why the Spark and HiveServer2 data access speeds are slow boil down to a couple of factors:
- Data is transferred in a form that is expensive to deserialize.
Data is passing through scalar Python objects (i.e. using
DataFrame.from_records on a list of tuples) rather than going directly into
pandas objects at the C API level.
Apache Arrow helps mitigate both of these problems. In the reasonably near future, I expect things architecturally to look like this:
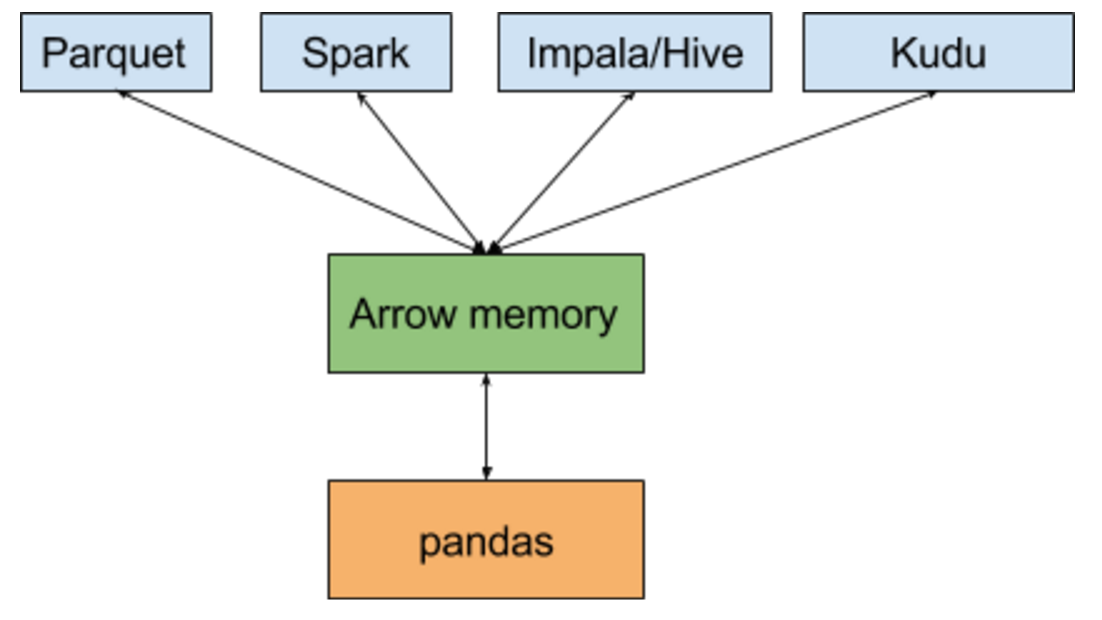
Realistically, the performance of ingesting data into pandas via Arrow should be significantly faster than reading a CSV (being binary and columnar).
It’s important to note that using pandas.read_csv as a standard for data
access performance doesn’t completely make sense. Parsing a CSV is fairly
expensive, which is why reading from HDF5 is 20x faster than parsing a CSV.
On Apache Parquet
The Apache Parquet data format is a column-oriented binary storage format for structured data optimized for IO throughput and fast analytics. Since it was designed primarily for use in a MapReduce setting initially, most development energy was poured into the parquet-mr Java implementation.
Last month I started getting involved in parquet-cpp, a native C++11 implementation of Parquet. I’m pleased to report we’ve made great progress on this in the last 6 weeks, and native read/write support for pandas users is reasonably near on the horizon. I’ll report here when I get the whole thing working end-to-end.
Summary and the road ahead
Data access performance is only one area where Arrow will help the Python ecosystem. High performance Python extensions and native in-memory handling for nested columnar data will also make a big impact. I look forward to sharing ongoing progress updates.